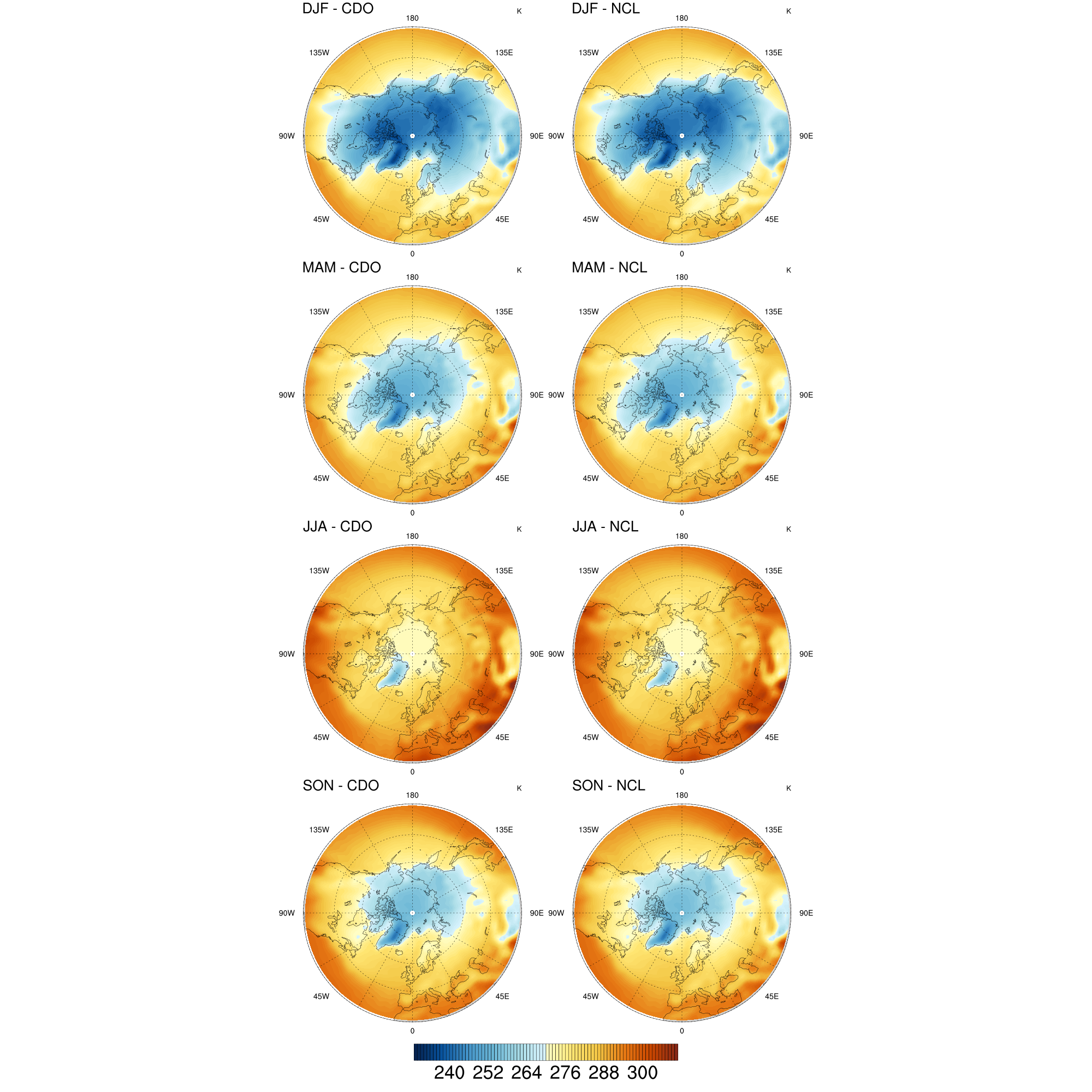
DKRZ NCL seasons comparison with CDO seasons#
Example script:
;--------------------------------------------------------------------
; DKSR NCL example: compute_seasons_compare_CDO_and_NCL.ncl
;
; Description: compare CDO and NCL seasons
;
; 08.06.18 kmf
;--------------------------------------------------------------------
begin
diri = "data/CMIP5/atmos/"
fili = "tas_Amon_MPI-ESM-LR_historical_r3i1p1_185001-200512.nc"
;-- open file and read variable
inf = addfile(diri+fili,"r")
var = inf->tas
;-- CDO compute seasons - compute "DJF","MAM","JJA","SON"
system("cdo -O -seasmean "+diri+fili+" cdo_outfile.nc")
f = addfile("cdo_outfile.nc","r")
var1c = f->tas(0::4,:,:)
var2c = f->tas(1::4,:,:)
var3c = f->tas(2::4,:,:)
var4c = f->tas(3::4,:,:)
;-- average over time
v1c = dim_avg_n_Wrap(var1c(:,{30:90},:),0)
v2c = dim_avg_n_Wrap(var2c(:,{30:90},:),0)
v3c = dim_avg_n_Wrap(var3c(:,{30:90},:),0)
v4c = dim_avg_n_Wrap(var4c(:,{30:90},:),0)
;-- NCL compute seasons - DJF, MAM, JJA, SON
seas = month_to_seasonN(var,(/"DJF","MAM","JJA","SON"/))
;-- average over time
v1n = dim_avg_n_Wrap(seas(0,:,{30:90},:),0)
v2n = dim_avg_n_Wrap(seas(1,:,{30:90},:),0)
v3n = dim_avg_n_Wrap(seas(2,:,{30:90},:),0)
v4n = dim_avg_n_Wrap(seas(3,:,{30:90},:),0)
;-- open workstation
wks_type = "png"
wks_type@wkWidth = 2000
wks_type@wkHeight = 2000
wks = gsn_open_wks(wks_type,"plot_seasmean_compare_CDO_and_NCL")
;-- set resources
res = True
res@gsnDraw = False
res@gsnFrame = False
res@gsnLeftString = ""
res@gsnLeftStringFontHeightF = 0.03
res@gsnPolar = "NH"
res@gsnPolarLabelDistance = 1.1 ;-- default dist. is 1.04
res@gsnPolarLabelFontHeightF = 0.015
res@gsnPolarLabelSpacing = 45.0 ;-- how frequently to label
res@gsnPolarLabelDistance = 1.08 ;-- default is 1.04
res@gsnPolarLabelFontHeightF = 0.025 ;-- change font height of labels
res@gsnPolarLabelFontHeightF = 0.016
res@mpMinLatF = 30 ;-- minimum lat to plot
res@mpCenterLonF = 0 ;-- center longitude
res@cnFillOn = True
res@cnLineLabelsOn = False
res@cnLinesOn = False
res@cnFillPalette = "BlueYellowRed"
res@cnLevelSelectionMode = "ManualLevels"
res@cnMinLevelValF = 230.0 ;-- contour min. value
res@cnMaxLevelValF = 310.0 ;-- contour max. value
res@cnLevelSpacingF = 1.0 ;-- contour interval
res@lbLabelBarOn = False
;-- create each single plot
res@gsnLeftString = "DJF - CDO"
plot1 = gsn_csm_contour_map_polar(wks,v1c,res)
res@gsnLeftString = "DJF - NCL"
plot2 = gsn_csm_contour_map_polar(wks,v1n,res)
res@gsnLeftString = "MAM - CDO"
plot3 = gsn_csm_contour_map_polar(wks,v2c,res)
res@gsnLeftString = "MAM - NCL"
plot4 = gsn_csm_contour_map_polar(wks,v2n,res)
res@gsnLeftString = "JJA - CDO"
plot5 = gsn_csm_contour_map_polar(wks,v3c,res)
res@gsnLeftString = "JJA - NCL"
plot6 = gsn_csm_contour_map_polar(wks,v3n,res)
res@gsnLeftString = "SON - CDO"
plot7 = gsn_csm_contour_map_polar(wks,v4c,res)
res@gsnLeftString = "SON - NCL"
plot8 = gsn_csm_contour_map_polar(wks,v4n,res)
;-- create the panel plot
pres = True
pres@gsnPanelLabelBar = True ;-- add common colorbar
gsn_panel(wks,(/plot1,plot2,plot3,plot4,plot5,plot6,plot7,plot8/),(/4,2/),pres)
end
Result: