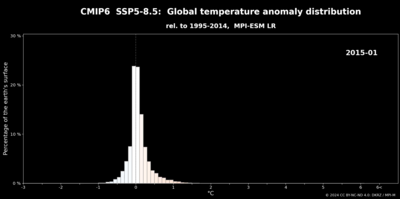
Python: global temperature anomaly distribution#
Description
In this example we visualize the shift in the global monthly temperature anomalies for the time period 2015-2100 relative to the simulated monthly temperatures in 1995-2014 based on the ensemble mean of the 10 CMIP6-realizations simulated for the pessimistic scenario SSP5-8.5 using the Earth system model MPI-ESM-LR.
Inspired by a nice visualization of the shifting distribution of temperature anomalies on land between 1963 and 2023, published by the NASA Scientific Visualization Studio <https://svs.gsfc.nasa.gov/5211/> based on the GISS surface temperature analysis (GISTEMP).
Software requirements
Python 3
matplotlib
xarray
numpy
python-cdo
Example script#
global_temperature_anomaly_distribution_animation_v5_1920x1080.py
#!/usr/bin/env python
# coding: utf-8
#------------------------------------------------------------------------------
# CMIP6: Global surface temperature anomaly distribution
#
#------------------------------------------------------------------------------
#
# global_temperature_anomaly_distribution_animation_v5_1920x1080.ipynb
#
# 2024 copyright DKRZ licensed under CC BY-NC-ND 4.0
# (https://creativecommons.org/licenses/by-nc-nd/4.0/deed.en)
#
#------------------------------------------------------------------------------
#
# Description
#
# Visualize the shift in the global monthly temperature anomalies for the time
# period 2015-2100 relative to the simulated monthly temperatures in 1995-2014
# based on the ensemble mean of the 10 CMIP6-realizations simulated for the
# pessimistic scenario SSP5-8.5 using the Earth system model MPI-ESM-LR.
#
# Data computations
#
# - The climatology was calculated from the ensemble mean of the historical data
# for the time period 1995-2015.
# - The anomalies were calculated by subtracting the climatology from the
# scenario SSP5-8.5 data.
# - These calculations were performed with CDO (Climate Data Operators,
# https://code.mpimet.mpg.de/projects/cdo).
#
# Data
#
# - CMIP6 MPI-ESM1.2-LR ensemble means
# - historical
# - SSP5-8.5
# - variable tas (surface temperature)
# - monthly means
#
# Inspired by
#
# Inspired by a nice visualization of the shifting distribution of temperature
# anomalies on land between 1963 and 2023, published by the Nasa Visualization
# Studio (https://svs.gsfc.nasa.gov/5211/) based on the GISS surface temperature
# analysis (GISTEMP).
#
#------------------------------------------------------------------------------
import os
import time
import functools
import numpy as np
import xarray as xr
import matplotlib.pyplot as plt
import matplotlib.cm as mcm
import matplotlib.colors as mcolors
from matplotlib.patches import Rectangle
from matplotlib import animation
from cdo import Cdo
cdo = Cdo()
# Data
#
#-- set the path to the CMIP6 MPI-ESM1.2-LR historical and SSP5-8.5 data
path = './data/CMIP6/LR/Temperature_Surface/'
hist_file = path + 'Historical_Runs/hist_em_LR_temp2_1995-2014.nc'
ssp_file = path + 'Runs/ssp585/ssp585_em_LR_temp_1995-2100.nc'
# Compute the anomalies
#-- compute the climatology
clim_file = cdo.ymonmean(input=hist_file)
#-- extract the time range 2015 to 2100 from the ssp5-8.5 input file
infile = cdo.selyear('2015/2100', input=ssp_file)
#-- compute the anomalies
anom_file = 'anom_ssp585.nc'
if os.path.exists(anom_file): os.remove(anom_file) #-- delete existing file
anom_file = cdo.sub(input=infile+' '+clim_file, output=anom_file, options='-O')
#-- open anomaly dataset and read variable tas
ds_anom = xr.open_dataset('anom_ssp585.nc')
tas = ds_anom.tas
# Set variables for weights computing.
NCELLS = tas.lat.size * tas.lon.size
EARTH_area = 510072000 #-- km**2
cell_area = EARTH_area / NCELLS
print('NCELLS = ', NCELLS)
# Settings
#
# Settings for the x- and y-axis as well as the time string.
#-- x-axis bounds
bndsmin = -3.
bndsmax = 6.
bndsinc = 0.1
nbounds = int((bndsmax + abs(bndsmin)) * 10)+1
bounds = np.linspace(bndsmin, bndsmax, nbounds)
npbounds = np.append(bounds, bounds[-1]+bndsinc)
x = bounds + bndsinc/2
nx = len(x)
#-- y-axis bounds
yaxismin = 0.
yaxismax = 5600
#-- time settings
times = ds_anom.time
time_str = ds_anom.time.dt.strftime('%Y-%m').data
# Check data and settings
#-- use time step
tstep = 0
weights = np.cos(np.deg2rad(ds_anom.lat))
with xr.set_options(keep_attrs=True):
data_weighted = (tas.isel(time=tstep) * weights).values.ravel()
#-- compute the histogram for the area weighted data
wn, wbins = np.histogram(data_weighted, npbounds)
# How many values are outside the last bin?
outer_list = np.empty(ds_anom.time.size)
for t in range(ds_anom.time.size):
dw = (tas.isel(time=t) * weights).values.ravel()
outer = np.array(dw)
outer[dw <= bndsmax] = 0
outer_list[t] = len(outer[outer != 0])
print(len(outer_list[outer_list != 0]))
print(min(outer_list), max(outer_list))
# Have a look at the bins.
#np.set_printoptions(formatter={'float': '{: 0.3f}'.format})
#print(f'Number of bins generated by numpy {len(wbins)}')
#print(wbins)
# Count negative and positive bins to be used to generate the colormap.
nneg = len(wbins[wbins<=(bndsinc*-1)])
npos = len(wbins[wbins>bndsinc])
nzero = len(wbins[wbins==0.])
print(nneg, npos, nzero)
# Colormap
#
# Generate an asymetric colormap with colors from dark blue to dark red with
# white in the middle (at x=zero).
#- generate the color arrays
whites = [[1., 1., 1., 1.]]*2
blues = mcm.Blues_r(np.linspace(0, 1, 128))
reds = mcm.Reds(np.linspace(0, 1, npos))
#-- choose the colors based on the number of bins for the negative or positive values
sel_blues = blues[len(blues)-nneg:]
sel_reds = reds[:npos]
#-- merge these color arrays and generate a colormap object
colors = np.vstack((sel_blues, whites, sel_reds))
cmap = mcolors.LinearSegmentedColormap.from_list('seismic_mod2', colors, N=len(colors))
# Get last color for the 'greater 6' bar.
dark_red = cmap(cmap.N)
print(dark_red)
# Test plot
plt.style.use('dark_background')
fig = plt.figure(figsize=(16, 9))
ax = fig.add_subplot()
fig.subplots_adjust(top=0.6) #-- keep space above plot
#-- title strings
tx_y = 0.82
plt.text(x=0.55, y=tx_y, s='CMIP6 SSP5-8.5: Global temperature anomaly distribution',
fontsize=24, ha='center', weight='bold', transform=fig.transFigure)
plt.text(x=0.55, y=tx_y-0.06, s= 'rel. to 1995-2014, MPI-ESM LR',
fontsize=18, weight='bold', ha='center', transform=fig.transFigure)
fig.text(0.81, 0.018, '© 2024 CC BY-NC-ND 4.0: DKRZ / MPI-M', fontsize=10)
#-- time string
tx = ax.text(5.6, 4800, time_str[0], fontsize=20, weight='bold')
#-- axis settings
ax.xaxis.label.set_size(12)
ax.yaxis.label.set_size(12)
ax.tick_params(axis='both', which='major', labelsize=12)
#-- x-tick values and labels
xtick_values = np.arange(bounds.min(), bounds.max()+1, 0.5)
ax.set_xticks(xtick_values)
xtick_labels = [ str(int(x)) for x in xtick_values ]
for i in range(len(xtick_values)):
if i % 2:
xtick_labels[i] = ' '
#-- add label for last xtick '6<'
xtick_labels[-1] = f'{int(bounds.max())}<'
ax.set_xticklabels(xtick_labels)
#-- y-tick values and labels
ax.set_yticks(ytick_values)
ax.set_yticklabels(ytick_labels)
#-- general axis settings
ax.set(xlim=[bounds.min(), bounds.max()+1],
ylim=[yaxismin, yaxismax],
xlabel='°C',
ylabel='%')
#-- draw vertical line at x = 0
zero = ax.axvline(x=0, ymin=yaxismin, ymax=yaxismax, color='gray', alpha=0.5, ls='--', lw=1.4)
#-- plot histogram time step tstep
plot2 = ax.hist(data_weighted,
bins=nx,
range=[bounds.min(),bounds.max()+bndsinc],
ec='grey',
lw=0.9,)
#-- add last bin to plot for values greater bndsmax
last_bar = ax.add_patch(Rectangle((bndsmax+0.45, 0.), 0.1, outer_list[tstep], facecolor=dark_red))
#-- draw line on top of the bars
line2, = ax.plot(x, plot2[0], lw=1.2, color='black', label='data weighted')
#-- set colors for bars
bin_centers2 = 0.5 * (plot2[1][:-1] + plot2[1][1:])
col2 = bin_centers2 - min(bin_centers2)
col2 /= max(col2)
for c, p in zip(col2, plot2[2]):
plt.setp(p, 'facecolor', cmap(c))
#-- adjust the padding between title and plot
fig.tight_layout()
#------------------------------------------------------------------------------
#
# Stop here if you do not want to generate the full animation
#
#------------------------------------------------------------------------------
exit()
#------------------------------------------------------------------------------
#
# Animation
#
# Function plot_init()
#
# Initilize some plot routines.
def plot_init(fig, ax):
tx_y = 0.82
plt.text(x=0.55, y=tx_y, s='CMIP6 SSP5-8.5: Global temperature anomaly distribution',
fontsize=24, weight='bold', ha='center', transform=fig.transFigure)
plt.text(x=0.55, y=tx_y-0.06, s= 'rel. to 1995-2014, MPI-ESM LR',
fontsize=18, weight='bold', ha='center', transform=fig.transFigure)
fig.text(0.81, 0.018, '© 2024 CC BY-NC-ND 4.0: DKRZ / MPI-M', fontsize=10)
# Function update_objects()
#
# Faster approach for animations. Only the given objects are changed so that the
# entire plot does not have to be regenerated for each frame.
def update_objects(frame, bounds, xcenter, bar_container, col):
with xr.set_options(keep_attrs=True):
data_weighted = (tas.isel(time=frame) * weights).values.ravel()
wn, wbins = np.histogram(data_weighted, npbounds)
for count, rect in zip(wn, bar_container):
rect.set_height(count)
for c, p in zip(col, bar_container):
plt.setp(p, 'facecolor', cmap(c))
#-- update the line object
#line2.set_xdata(xcenter)
#line2.set_ydata(wn)
last_bar.set_height(outer_list[frame])
#-- update date string time_str object
tx.set_text(time_str[frame])
#return (bar_container.patches, line2, tx, last_bar)
return (bar_container.patches, tx, last_bar)
# Number of total frames (time steps)
frames = times.size
# ffmpeg binary path
#
# Use a newer ffmpeg version from a cartopy environment. If you have a new
# ffmpeg version in your environment modify the lines below accordingly.
ffmpeg_path = os.environ['HOME']+'/miniconda3/envs/cartopy/bin/ffmpeg'
plt.rcParams['animation.ffmpeg_path'] = ffmpeg_path
# Initialize the data
weights = np.cos(np.deg2rad(ds_anom.lat))
with xr.set_options(keep_attrs=True):
data_weighted = (tas.isel(time=0) * weights).values.ravel()
#-- compute the histogram for the area weighted data
wn, wbins = np.histogram(data_weighted, npbounds)
# Generate the animation
#
# Run times: writer dpi=100 220 s
# writer dpi=150 330 s
# figure dpi=1920/16 102 s optimized
#frames = 10
MPEG_FILE = 'global_temperature_distribution_animation_v5_1920x1080.mp4'
#-----------------------------------------------------------------------
t1 = time.time()
plt.style.use('dark_background')
#------------------------------------
#-- create a figure and add subplot
#------------------------------------
fig = plt.figure(figsize=(16, 9), dpi=1920/16) #-- 1920x1080
ax = fig.add_subplot()
fig.subplots_adjust(top=0.6) #-- keep space above plot for the title strings
#------------------------------------
#-- axis settings
#------------------------------------
ax.xaxis.label.set_size(12)
ax.yaxis.label.set_size(12)
ax.tick_params(axis='both', which='major', labelsize=12)
xtick_values = np.arange(bounds.min(), bounds.max()+1, 0.5)
ax.set_xticks(xtick_values)
#-- draw every other xtick label
xtick_labels = [ str(int(x)) for x in xtick_values ]
for i in range(len(xtick_values)):
if i % 2:
xtick_labels[i] = ' '
#-- add label for last xtick '6<'
xtick_labels[-1] = f'{int(bounds.max())}<'
ax.set_xticklabels(xtick_labels)
#-- y-tick values and labels
ytick_values = [ (NCELLS*x)/100 for x in range(0,110,10)]
ytick_labels = [ f'{x} %' for x in range(0,110,10)]
ax.set_yticks(ytick_values)
ax.set_yticklabels(ytick_labels)
#-- general axis settings
ax.set(xlim=[bounds.min(), bounds.max()+1],
ylim=[yaxismin, yaxismax],
xlabel='°C',
ylabel='')
#------------------------------------
#-- init title to fix the figure size
#------------------------------------
tx_y = 0.82
plt.text(x=0.55, y=tx_y, s='CMIP6 SSP5-8.5: Global temperature anomaly distribution',
fontsize=24, ha='center', weight='bold', transform=fig.transFigure)
plt.text(x=0.55, y=tx_y-0.06, s= 'rel. to 1995-2014, MPI-ESM LR',
fontsize=18, weight='bold', ha='center', transform=fig.transFigure)
#------------------------------------
#-- add date string to plot
#------------------------------------
tx = ax.text(5.6, 4800, time_str[0], fontsize=20, weight='bold')
#------------------------------------
#-- draw vertical line at x = 0
#------------------------------------
zero = ax.axvline(x=0, ymin=yaxismin, ymax=yaxismax, color='gray', alpha=0.5,
ls='--', lw=1.4)
#------------------------------------
#-- plot histogram time step tstep
#------------------------------------
plot2 = ax.hist(data_weighted,
bins=nx,
range=[bounds.min(),bounds.max()+bndsinc],
ec='grey',
lw=0.9,)
#---------------------------------------------------
#-- add last bin to plot for values greater bndsmax
#--------------------------------------------------
last_bar = ax.add_patch(Rectangle((bndsmax+0.45, 0.), 0.1, outer_list[tstep],
facecolor=dark_red))
#---------------------------------
#-- draw hist data as line
#---------------------------------
#line2, = ax.plot(x, plot2[0], lw=1.2, color='black', label='data weighted')
#---------------------------------
#-- set colors for bars
#---------------------------------
bin_centers = 0.5 * (plot2[1][:-1] + plot2[1][1:])
col = bin_centers - min(bin_centers)
col /= max(col)
for c, p in zip(col, plot2[2]):
plt.setp(p, 'facecolor', cmap(c))
#-- adjust the padding between title and plot
fig.tight_layout()
#------------------------------------
#-- animation
#------------------------------------
#-- create partial callable objects
#------------------------------------
pltinit = functools.partial(plot_init, fig=fig, ax=ax)
update = functools.partial(update_objects,
col=col,
bounds=bounds,
xcenter=x,
bar_container=plot2[2])
#-- generate animation (increase 'interval' to slow down the animation)
ani = animation.FuncAnimation(fig=fig,
func=update,
init_func=pltinit,
frames=range(0, frames),
interval=2000)
#-- save the animation to MPEG file
writer = animation.FFMpegWriter(fps=15, bitrate=-1, codec='h264')
ani.save(MPEG_FILE, writer=writer)
#-- close figure window
plt.close()
#-- print run time
t2 = time.time()
print(f'Run time for # {frames} frames: {(t2-t1):.1f} s')