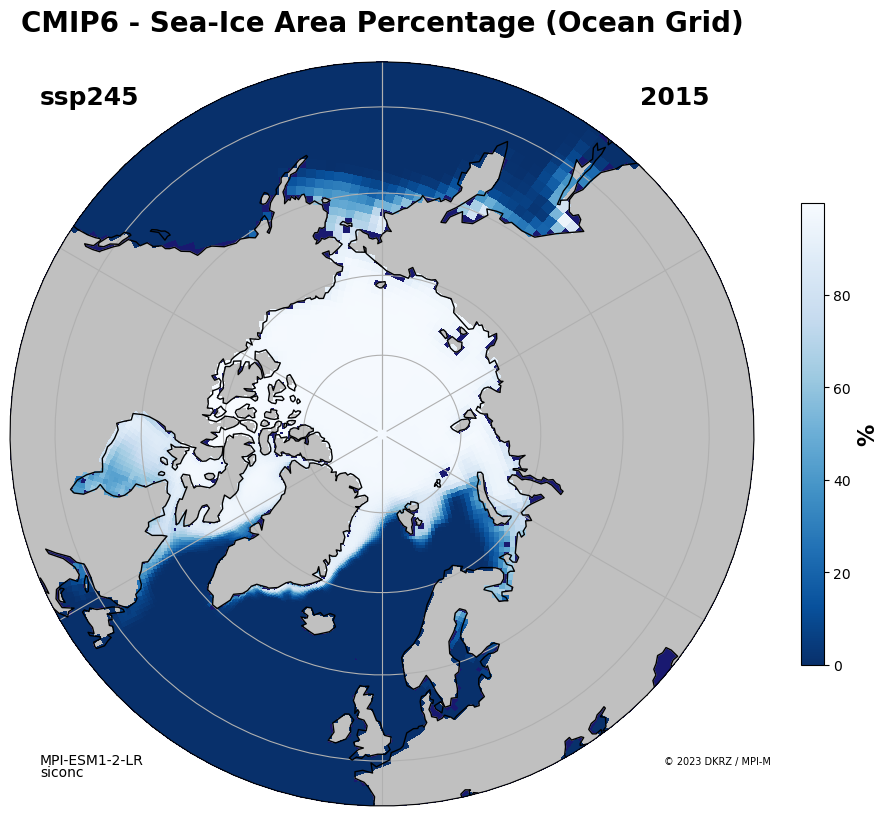
Python sea ice polar plot#
Software requirements:
Python 3
numpy
xarray
matplotlib
cartopy
Example script#
sea_ice_thick_conc_mass_CMIP6.py
#!/usr/bin/env python
# coding: utf-8
'''
DKRZ example
Sea ice data on polar plot
Create a map plot of the sea ice data with North-Polar_Stereographic projection.
Variable siconc: Sea-Ice Area Percentage (Ocean Grid)
-------------------------------------------------------------------------------
2023 copyright DKRZ licensed under CC BY-NC-SA 4.0
(https://creativecommons.org/licenses/by-nc-sa/4.0/deed.en)
-------------------------------------------------------------------------------
'''
import os
import xarray as xr
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.path as mpath
import cartopy.crs as ccrs
import cartopy.feature as cfeature
# Fix the wrapped lines problem
#
# Function z_masked_overlap()
#
# The function z_masked_overlap was taken from
# https://github.com/SciTools/cartopy/issues/1421 from htonchia
def z_masked_overlap(axe, X, Y, Z, source_projection=None):
'''
For data in projection axe.projection
find and mask the overlaps (more 1/2 the axe.projection range)
X, Y either the coordinates in axe.projection or longitudes latitudes
Z the data
operation one of 'pcorlor', 'pcolormesh', 'countour', 'countourf'
if source_projection is a geodetic CRS data is in geodetic coordinates
and should first be projected in axe.projection
X, Y are 2D same dimension as Z for contour and contourf
same dimension as Z or with an extra row and column for pcolor
and pcolormesh
return ptx, pty, Z
'''
if not hasattr(axe, 'projection'):
return X, Y, Z
if not isinstance(axe.projection, ccrs.Projection):
return X, Y, Z
if len(X.shape) != 2 or len(Y.shape) != 2:
return X, Y, Z
if (source_projection is not None and
isinstance(source_projection, ccrs.Geodetic)):
transformed_pts = axe.projection.transform_points(
source_projection, X, Y)
ptx, pty = transformed_pts[..., 0], transformed_pts[..., 1]
else:
ptx, pty = X, Y
with np.errstate(invalid='ignore'):
# diagonals have one less row and one less columns
diagonal0_lengths = np.hypot(
ptx[1:, 1:] - ptx[:-1, :-1],
pty[1:, 1:] - pty[:-1, :-1])
diagonal1_lengths = np.hypot(
ptx[1:, :-1] - ptx[:-1, 1:],
pty[1:, :-1] - pty[:-1, 1:])
to_mask = ((diagonal0_lengths > (
abs(axe.projection.x_limits[1]
- axe.projection.x_limits[0])) / 2) |
np.isnan(diagonal0_lengths) |
(diagonal1_lengths > (
abs(axe.projection.x_limits[1]
- axe.projection.x_limits[0])) / 2) |
np.isnan(diagonal1_lengths))
# TODO check if we need to do something about surrounding vertices
# add one extra colum and row for contour and contourf
if (to_mask.shape[0] == Z.shape[0] - 1 and
to_mask.shape[1] == Z.shape[1] - 1):
to_mask_extended = np.zeros(Z.shape, dtype=bool)
to_mask_extended[:-1, :-1] = to_mask
to_mask_extended[-1, :] = to_mask_extended[-2, :]
to_mask_extended[:, -1] = to_mask_extended[:, -2]
to_mask = to_mask_extended
if np.any(to_mask):
Z_mask = getattr(Z, 'mask', None)
to_mask = to_mask if Z_mask is None else to_mask | Z_mask
Z = np.ma.masked_where(to_mask, Z)
return ptx, pty, Z
def main():
plt.switch_backend('agg')
# LR Scenario data
ddir = os.environ['HOME']+'/data/CMIP6/LR/SeaIce/Runs/'
ssps = ['ssp126', 'ssp245', 'ssp370','ssp585']
fsiconc = 'LR_SImon_EM_SICONC.nc'
scen_name = ssps[1]
path_to_data = ddir + scen_name + '/' + fsiconc
# Read data
ds = xr.open_dataset(path_to_data)
var = ds.siconc.isel(time=0)
lon = ds.longitude
lat = ds.latitude
# Select a time step, retrieve the date string
timestep = -1
datestep = var.time.dt.year.data
# Create pcolormesh plot
#
# Note:
# 1. use the ccrs.Stereographic projection for the transform setting
# 2. set a circular boundary for the plot
proj = ccrs.NorthPolarStereo()
kwtrans = dict(central_latitude=90, central_longitude=0.)
trans = ccrs.Stereographic(**kwtrans)
fig, ax = plt.subplots(figsize=(12,12), subplot_kw={"projection":proj})
X, Y, maskedZ = z_masked_overlap(ax, lon.data, lat.data, var,
source_projection=ccrs.Geodetic())
ax.set_extent([-180, 180, 45, 90], ccrs.PlateCarree())
ax.add_feature(cfeature.OCEAN, color='midnightblue', zorder=0)
ax.add_feature(cfeature.LAND, color='silver', zorder=1)
ax.add_feature(cfeature.COASTLINE, zorder=3)
ax.gridlines()
ax.set_title('CMIP6 - '+var.attrs['long_name'], y=1.03, fontsize=20, weight='bold')
plot = ax.pcolormesh(X, Y, maskedZ,
cmap='Blues_r',
transform=trans,
zorder=2)
cbar = plt.colorbar(plot, ax=ax, shrink=0.5, pad=0.05)
cbar.set_label(label=var.attrs['units'], size=16, weight='bold')
# Circular boundary of the map
# (see https://scitools.org.uk/cartopy/docs/v0.15/examples/always_circular_stereo.html)
theta = np.linspace(0, 2*np.pi, 100)
center = [0.5, 0.5]
radius = 0.5
verts = np.vstack([np.sin(theta), np.cos(theta)]).T
circle = mpath.Path(verts * radius + center)
ax.set_boundary(circle, transform=ax.transAxes)
fig.text(0.15, 0.77, scen_name, weight='bold', fontsize=18)
fig.text(0.65, 0.77, str(datestep), fontsize=18, weight='bold')
fig.text(0.15, 0.22, 'MPI-ESM1-2-LR', fontsize=10)
fig.text(0.15, 0.21, 'siconc', fontsize=10)
fig.text(0.67, 0.22, '© 2023 DKRZ / MPI-M', fontsize=7)
fig.savefig('plot_sea_ice_thick_conc_volume_'+scen_name+'.png',
bbox_inches='tight', facecolor='white', dpi=100)
if __name__ == '__main__':
main()