Posts by Sofiane Bendoukha
Container mode now available in the advanced spawner
- 01 July 2025
We’re excited to announce that container mode is now enabled in our advanced JupyterHub spawner.
Container mode allows users to choose from a list of pre-defined container images when starting a Jupyter session. Each image is tailored for specific workflows (e.g. data science, coding).
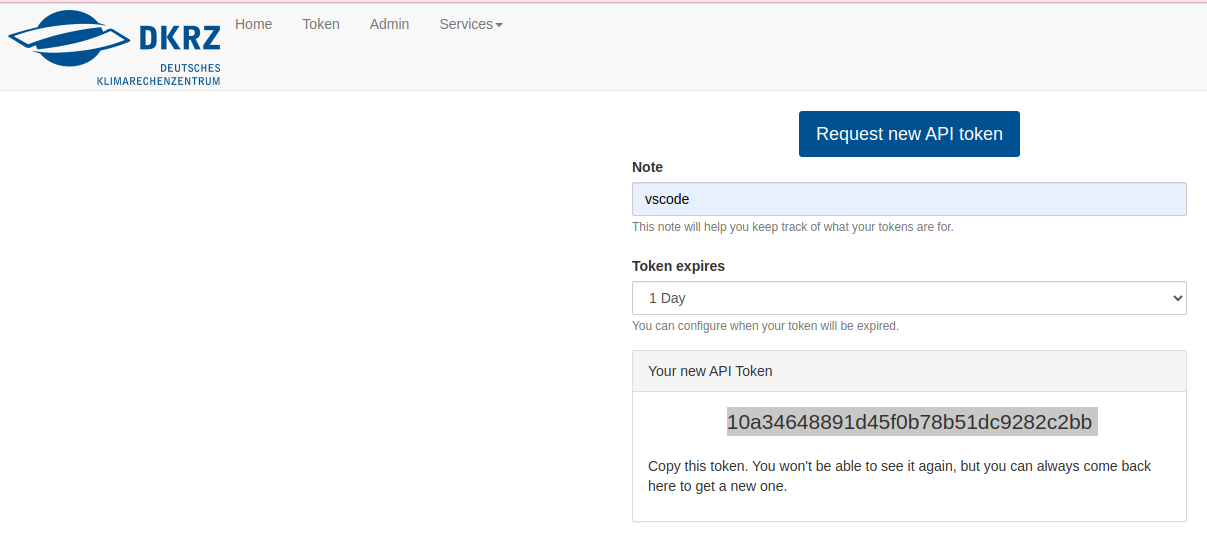
Set up Remote Developement/Debugging
- 24 February 2023
Some workarounds related to remote development/debugging on Levante. These workarounds are not tested on all available remote tools but often specific to a certain IDE. We encourage you to test on your favorite IDE and report issues to support@dkrz.de.
Tested on VSCode
How to install R packages in different locations?
- 25 October 2021
The default location for R packages is not writable and you can not install new packages. On demand we install new packages system-wide and for all users. However, it possible to install packages in different locations than root and here are the steps:
create a directory in $HOME e.g. ~/R/libs
JupyterDash on Jupyterhub @ DKRZ
- 24 October 2021
As of Dash v2.11, Jupyter support is built into the main Dash package, which is installed in jupyterhub. You don’t need the jupyter_dash anymore. See https://dash.plotly.com/dash-in-jupyter for usage details.
This content is based on this notebook.
How to install jupyter kernel for Matlab
- 10 June 2021
Fixed broken link in Matlab release section.
In this tutorial, we will describe i) the steps to create a kernel for Matlab and ii) get the matlab_kernel working in Jupyterhub on Levante.
How to re-enable the deprecated python kernels?
- 16 May 2021
Within the maintenance of Monday, May 15th, we will perform updates in our python installations (please see the details here).
Since the jupyterhub kernels are based on modules, the deprecated kernels will no longer be available as default kernels in jupyter notebooks/labs.
Requested MovieWriter (ffmpeg) not available
- 06 May 2021
Requested MovieWriter (ffmpeg) not available
conda env with ffmpeg and ipykernel
Create a kernel from your own Julia installation
- 23 March 2021
We already provide a kernel for Julia based on the module julia/1.7.0.
In order to use it, you only need to install ÌJulia:
Python environment locations
- 04 March 2021
Kernels are based on python environments created with conda,
virtualenv or other package manager. In some cases, the size of the
environment can tremendously grow depending on the installed packages.
The default location for python files is the $HOME directory. In
this case, it will quickly fill your quota. In order to avoid this, we
suggest that you create/store python files in other directories of the
filesystem on Levante.
The following are two alternative locations where you can create your Python environment:
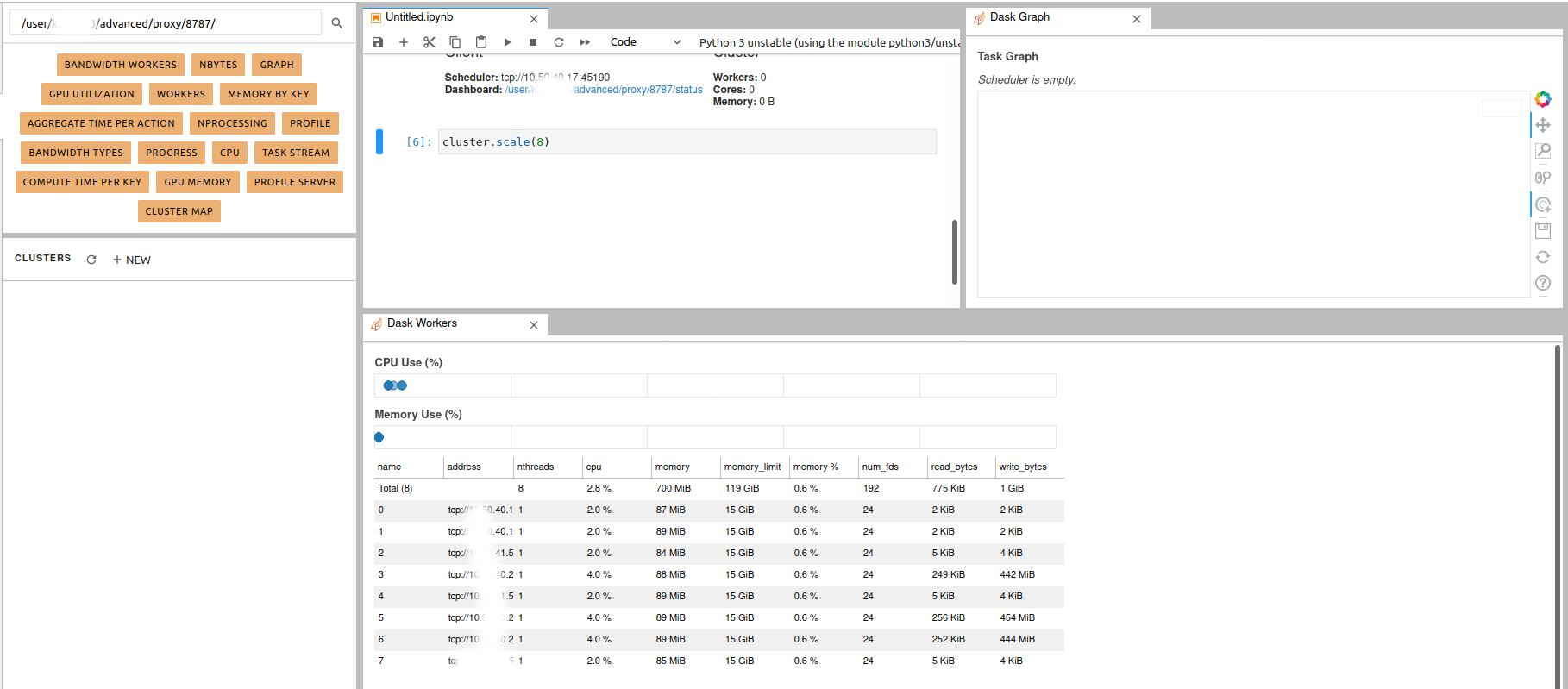
Dask jobqueue on Levante
- 16 November 2020
According to the official Web site, Dask jobqueue can be used to
deploy Dask on job queuing systems like PBS, Slurm, MOAB, SGE,
LSF, and HTCondor. Since the queuing system on Levante is Slurm, we are
going to show how to start a Dask cluster there. The idea is simple as
described here. The difference is that the workers can be distributed
through multiple nodes from the same partition. Using Dask jobqueue you can launch
Dask cluster/workers as a Slurm jobs. In this case, Jupyterhub will play an interface role and the Dask
can use more than the allocated resources to your jupyterhub session
(profiles).
Load the required clients
Enable NCL Kernel in Jupyterhub
- 05 November 2020
Can’t use NCL (Python) as kernel in Jupyter
This tutorial won’t work
FileNotFoundError: [Errno 2] No such file or directory
- 07 October 2020
See Wrapper packages here.
you:
Jupyterhub log file
- 25 September 2020
Each Jupyter notebook is running as a SLUM job on Levante. By default,
stdout and stderr of the SLURM batch job that is spawned by
Jupyterhub is written to your HOME directory on the HPC system. In
order to make it simple to locate the log file:
if you use the preset options form: the log file is named
jupyterhub_slurmspawner_preset_<id>.log.
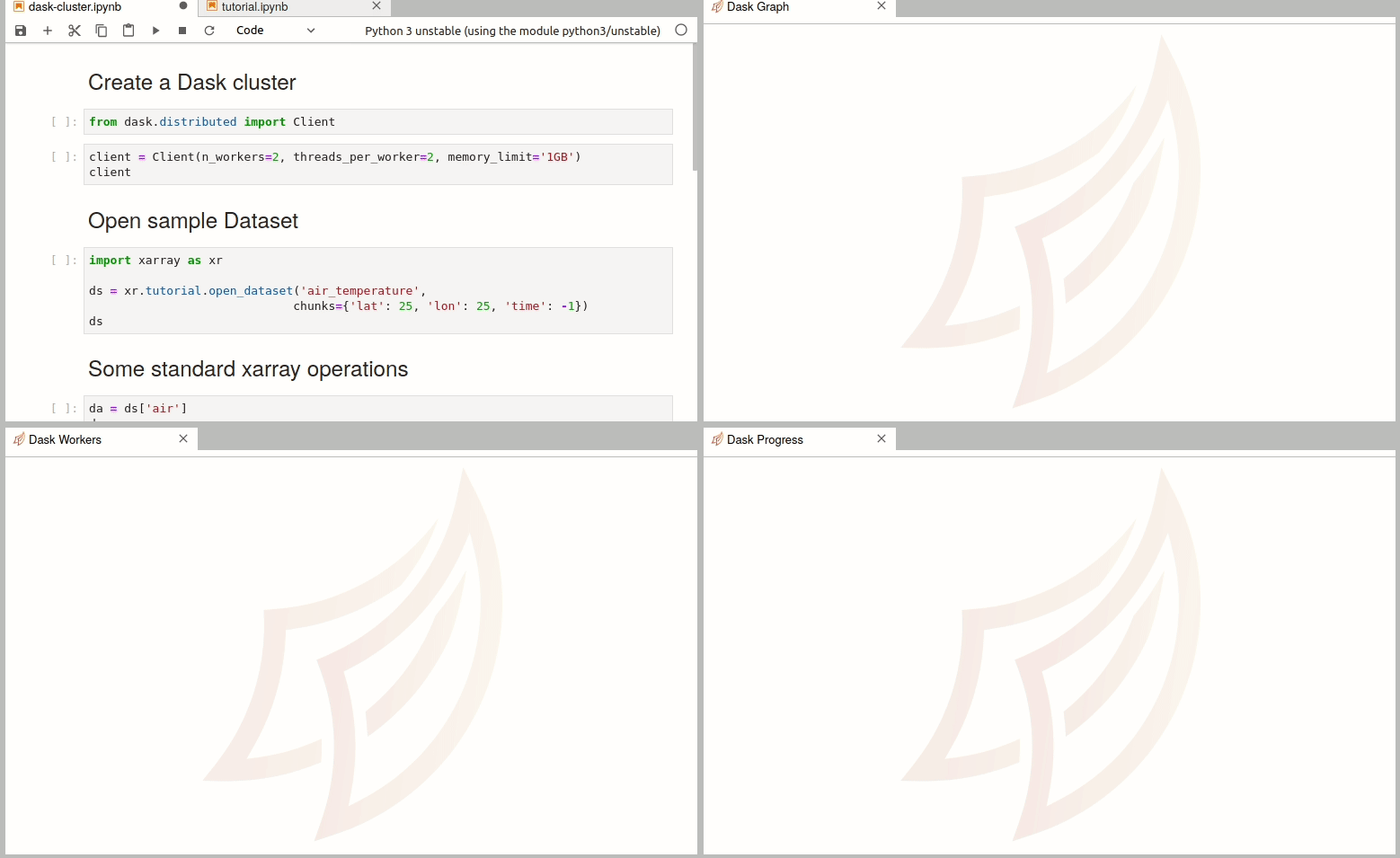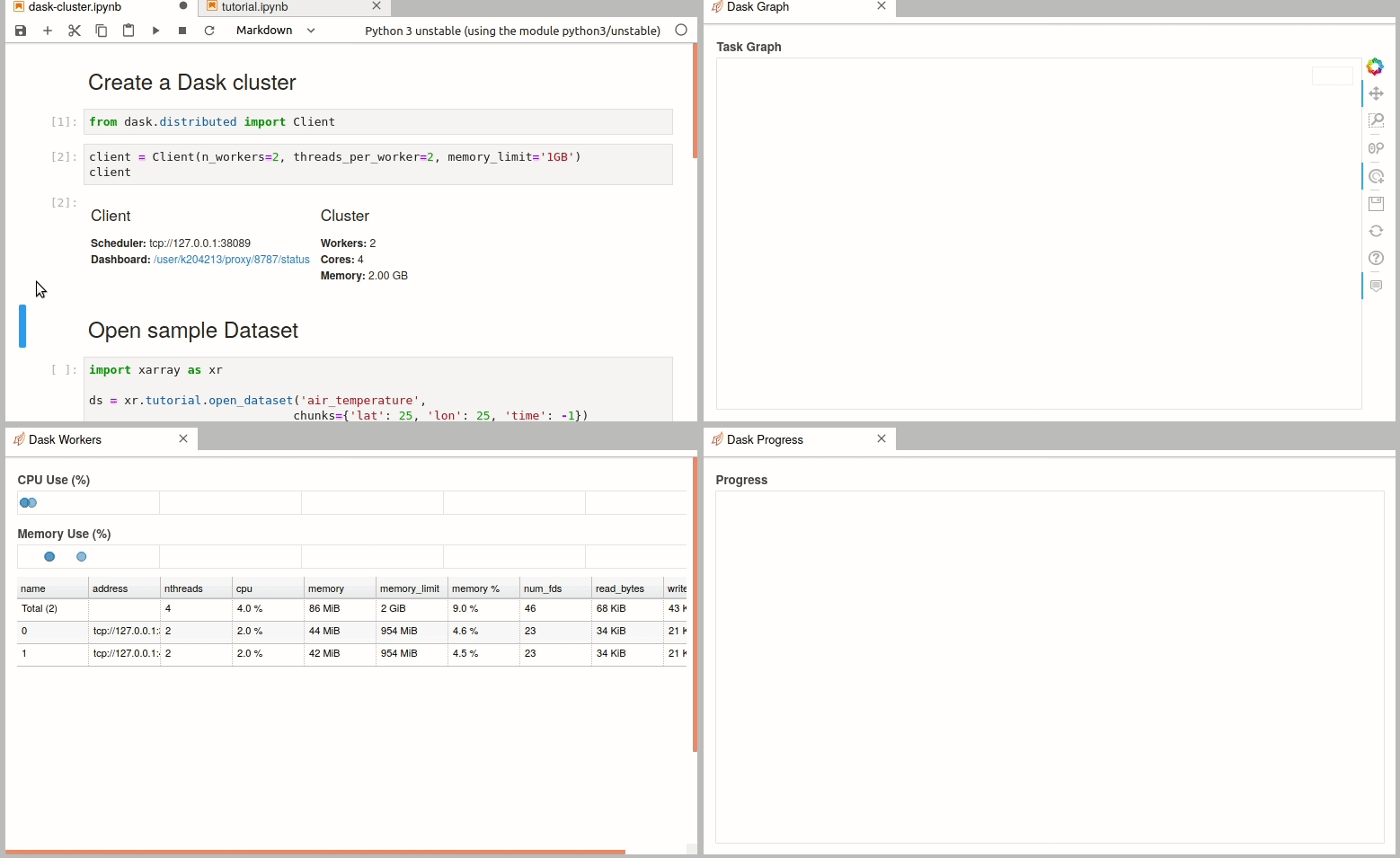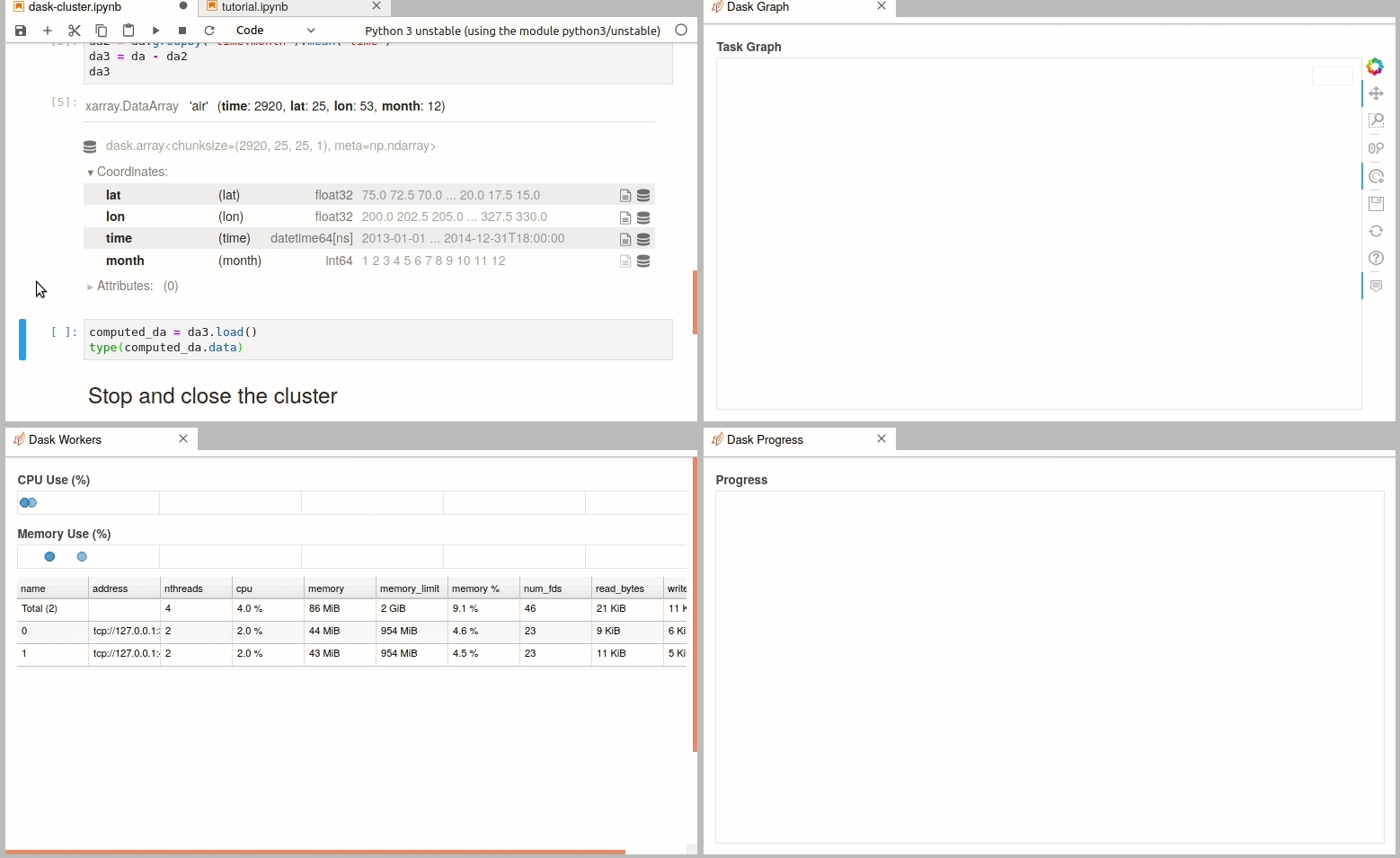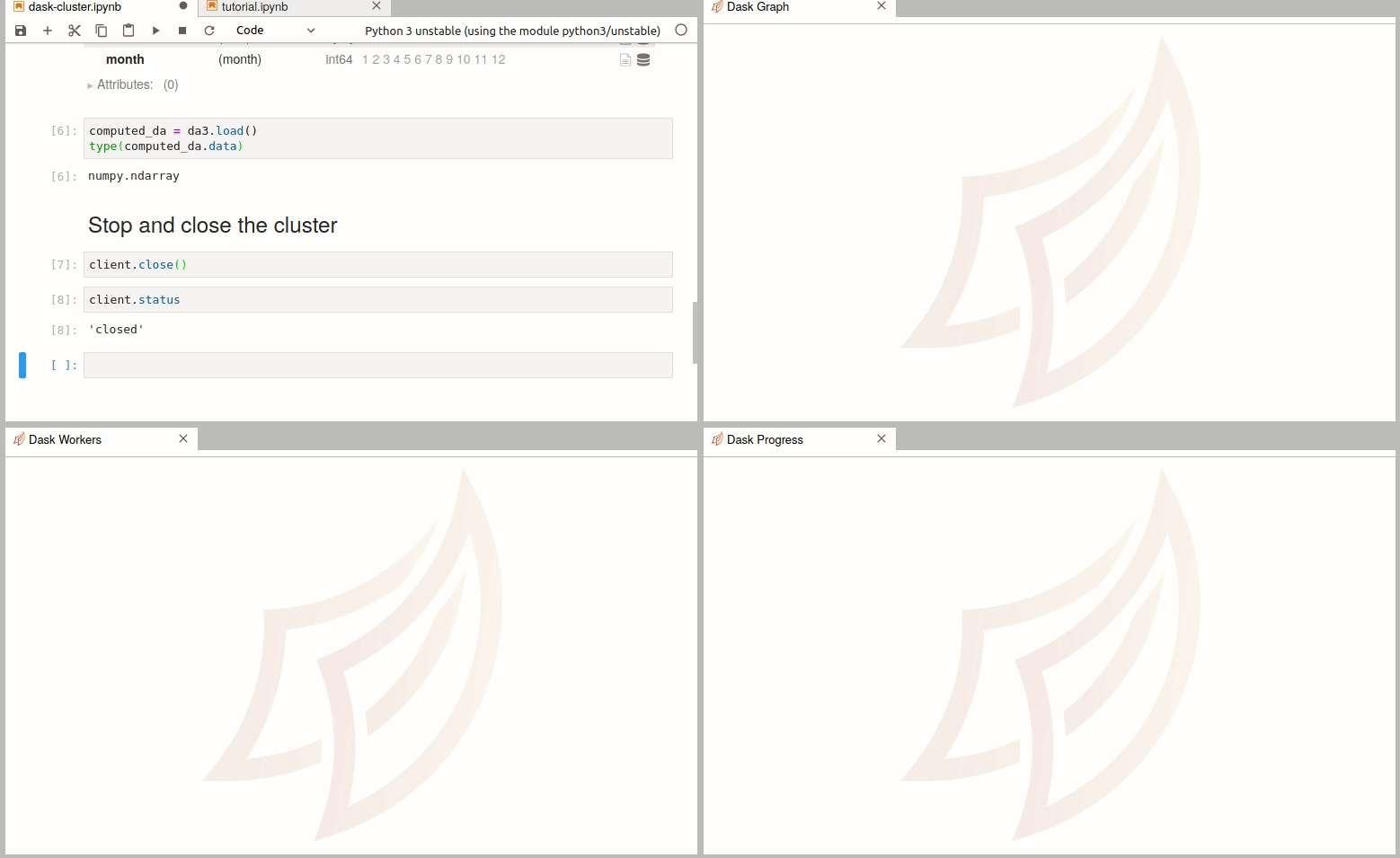
Simple Dask clusters in Jupyterhub
- 18 September 2020
There are multiple ways to create a dask cluster, the following is only an example. Please consult the official documentation. The Dask library is installed and can be found in any of the python3 kernels in jupyterhub. Of course, you can use your own python environment.
The simplest way to create a Dask cluster is to use the distributed module:
DKRZ Tech Talks
- 03 September 2020
It is our great pleasure to introduce the DKRZ Tech Talks. In this series of virtual talks we will present services of DKRZ and provide a forum for questions and answers. They will cover technical aspects of the use of our compute systems as well as procedures such as compute time applications and different teams relevant to DKRZ such as our machine learning specialists. The talks will be recorded and uploaded afterwards for further reference.
Go here for more information.