Databrowser#
All files available on Freva are scanned and indexed in a special server (SOLR). This allows us to query the server which responds almost immediately. Because of the Freva configuration the first time you call the tool it might take up to a couple of seconds to start. After that normally you should see results within a second.
The draw back of such a system is that the metadata regarding the files is detached from the file system where they are being held. This means that you might get files that are not there anymore or don’t get some that are indeed there. Normally files does not change that fast and you should see everything in there.
You might also use this command to get much more data than just a list of paths. Please see the use cases for a fast introduction to what can be done.
Help#
$ freva --databrowser --help
The query is of the form key=value. <value> might use *, ? as wildcards or any regular expression enclosed in forward slashes. Depending on your shell and the symbols used, remember to escape the sequences properly.
The safest would be to enclosed those in single quotes.
For Example:
%s project=baseline1 model=MPI-ESM-LR experiment=/decadal200[0-3]/ time_frequency=*hr variable='/ta|tas|vu/'
Usage: freva --databrowser [options]
Options:
-d, --debug turn on debugging info and show stack trace on
exceptions.
-h, --help show this help message and exit
--multiversion select not only the latest version but all of them
--relevant-only show only facets that filter results (i.e. >1 possible
values)
--batch-size=N Number of files to retrieve
--count-facet-values Show the number of files for each values in each facet
--attributes retrieve all possible attributes for the current
search instead of the files
--all-facets retrieve all facets (attributes & values) instead of
the files
--facet=FACET retrieve these facets (attributes & values) instead of
the files
Usage in CLI#
The freva --databrowser command expects a list of
attribute=value (or key=value) pairs. There are a few
differences and many more options (explained next). The most important
thing is that you don’t need to split the search according to the type
of data you are searching for. You might as well search for files both
on observations, reanalysis and model data all at the same time.
Also important is that all searches are made case insensitive (so don’t worry about upper or lower casing)
You can also search for attributes themselves instead of file paths. For example you can search for the list of variables available that satisfies a certain constraint (e.g. sampled 6hr, from a certain model, etc).
The search is defined in the following terms:
$ freva --databrowser project=baseline* variable=tas time_frequency=mon
There are 12 different attributes to search for. You can see them if
you double <TAB> after calling the databrowser (this feature works
under BASH, for more info go to bash auto
completion):
$ freva --databrowser <TAB><TAB>
cmor_table= ensemble= grid= model= project= time_frequency=
data_type= experiment= institute= product= realm= variable=
1. Defining the possible values#
There are many more options for defining a value for a given attribute:
Attribute syntax |
Meaning |
|---|---|
|
Search for files containing exactly that attribute |
|
Search for files containing a value for attribute that starts with the prefix |
|
Search for files containing a value for attribute that ends with the suffix |
|
Search for files containing a value for attribute that has |
|
Search for files containing a value for attribute that matches the given regular expression (check this table for some common regular expressions) |
|
Search for files containing either value1 OR value2 for the given attribute (note that’s the same attribute twice!) |
|
Search for files containing value1 for attribute1 AND value2 for attribute2 |
|
Search for files NOT containing value |
|
Search for files containin NEITHER value1 or value2 |
Note
When using * remember that your shell might give it a different meaning, normally it will try to match files with that name.
To turn that off you can use backslash \ in most shells (\*).
For more in to that go to the commonly used regular expressions list.
2. Searching for metadata#
You might as well want to now about possible values that an attribute
can take after a certain search is done. For this you use the
--facet flag (facets are the possible attributes that partition the
result set).
For example to see the time frequency (time resolution) in which reanalysis are available you might issue the following query:
$ freva --databrowser data_type=reanalysis --facet time_frequency
time_frequency: 1hr,3hr,6hr,amon,ann,day,fx,mon,yr
or
$ freva --databrowser --facet time_frequency data_type=reanalysis
time_frequency: 1hr,3hr,6hr,amon,ann,day,fx,mon,yr
Also note that you can further define this as usual with a given query. For example check which files are at 6hr frequency:
$ freva --databrowser --facet variable data_type=reanalysis time_frequency=6hr
variable:cape,cin,clh,clivi,cll,clm,clt,clwvi,hur1000,hur250,hur500,hur700,hur850,hur925,hurs,hus1000,hus250,hus500,hus700,hus850,hus925,pr,prhmax,prw,psl,sfcwind,sfcwindmax,sli,ta,ta1000,ta250,ta500,ta700,ta850,ta925,tas,tasmax,tasmin,td2m,tos,ua,ua1000,ua250,ua500,ua700,ua850,ua925,uas,va,va1000,va250,va500,va700,va850,va925,vas,wap,zg,zg1000,zg250,zg500,zg700,zg850,zg925
If you want to see how many files would return if you further select
that variable (drill down query) you may add the
--count-facet-values flag (simply --count will also do):
$ freva --databrowser --count-facet-values --facet variable data_type=reanalysis time_frequency=6hr
variable: cape (1),cin (1),clh (1),clivi (1),cll (1),clm (1),clt (1),clwvi (1),hur1000 (1),hur250 (1),hur500 (1),hur700 (1),hur850 (1),hur925 (1),hurs (1),hus1000 (1),hus250 (1),hus500 (1),hus700 (1),hus850 (1),hus925 (1),pr (34),prhmax (34),prw (1),psl (8169),sfcwind (1),sfcwindmax (34),sli (1),ta (238),ta1000 (1),ta250 (1),ta500 (1),ta700 (1),ta850 (1),ta925 (1),tas (101),tasmax (34),tasmin (34),td2m (1),tos (34),ua (143),ua1000 (1),ua250 (1),ua500 (1),ua700 (1),ua850 (1),ua925 (1),uas (7922),va (143),va1000 (1),va250 (1),va500 (1),va700 (1),va850 (1),va925 (1),vas (7922),wap (139),zg (383),zg1000 (1),zg250 (1),zg500 (1),zg700 (1),zg850 (1),zg925 (1)
This means that there are 8169 files containing the variable psl, 7922 files containing the variables vas and uas, and so on.
Currently there is no possibility to show the values of more than one attribute at a time (i.e. --facet variable --facet data_type or --facet variable,data_type does not work). However, you can check ALL facets at once. For that you may use the --all-facets
flag:
$ freva --databrowser --all-facets data_type=reanalysis time_frequency=6hr cmor_table=6hrplev
cmor_table: 6hrplev
product: reanalysis
v\ersion:
data_type: reanalysis
institute: ecmwf,jma-criepi,nasa-gmao,ncep-ncar,noaa-cires
project: reanalysis
time_frequency: 6hr
experiment: 20cr,20cr-em,cfsr,era40,era40-hr,eraint,jra-25,jra-25-fc,merra,ncep1,ncep2
grid:
variable: pr,prhmax,psl,sfcwindmax,ta,tas,tasmax,tasmin,tos,ua,uas,va,vas,wap,zg
realm: atmos,ocean
model: cdas,cfs,geos-5,gfs,ifs,jcdas
ensemble: r10i1p1,r11i1p1,r12i1p1,r13i1p1,r14i1p1,r15i1p1,r16i1p1,r17i1p1,r18i1p1,r19i1p1,r1i1p1,r20i1p1,r21i1p1,r22i1p1,r23i1p1,r24i1p1,r25i1p1,r26i1p1,r27i1p1,r28i1p1,r29i1p1,r2i1p1,r30i1p1,r31i1p1,r32i1p1,r33i1p1,r34i1p1,r35i1p1,r36i1p1,r37i1p1,r38i1p1,r39i1p1,r3i1p1,r40i1p1,r41i1p1,r42i1p1,r43i1p1,r44i1p1,r45i1p1,r46i1p1,r47i1p1,r48i1p1,r49i1p1,r4i1p1,r50i1p1,r51i1p1,r52i1p1,r53i1p1,r54i1p1,r55i1p1,r56i1p1,r5i1p1,r6i1p1,r7i1p1,r8i1p1,r9i1p1
And again you can also have the --count flag:
$ freva --databrowser --all-facets --count data_type=reanalysis time_frequency=6hr
cmor_table: 6hr (51),6hrplev (25362)
product: reanalysis (25413)
v\ersion:
data_type: reanalysis (25413)
institute: ecmwf (693),jma-criepi (336),miklip-module-c (51),nasa-gmao (101),ncep-ncar (520),noaa-cires (23712)
project: reanalysis (25413)
time_frequency: 6hr (25413)
experiment: 20cr (23520),20cr-em (192),cfsr (96),era40 (200),era40-hr (126),eraint (367),eraint-eur-22-cclm (51),jra-25 (132),jra-25-fc (204),merra (101),ncep1 (390),ncep2 (34)
grid:
variable: cape (1),cin (1),clh (1),clivi (1),cll (1),clm (1),clt (1),clwvi (1),hur1000 (1),hur250 (1),hur500 (1),hur700 (1),hur850 (1),hur925 (1),hurs (1),hus1000 (1),hus250 (1),hus500 (1),hus700 (1),hus850 (1),hus925 (1),pr (34),prhmax (34),prw (1),psl (8169),sfcwind (1),sfcwindmax (34),sli (1),ta (238),ta1000 (1),ta250 (1),ta500 (1),ta700 (1),ta850 (1),ta925 (1),tas (101),tasmax (34),tasmin (34),td2m (1),tos (34),ua (143),ua1000 (1),ua250 (1),ua500 (1),ua700 (1),ua850 (1),ua925 (1),uas (7922),va (143),va1000 (1),va250 (1),va500 (1),va700 (1),va850 (1),va925 (1),vas (7922),wap (139),zg (383),zg1000 (1),zg250 (1),zg500 (1),zg700 (1),zg850 (1),zg925 (1)
realm: atmos (25379),ocean (34)
model: cclm-eur-22 (51),cdas (424),cfs (96),geos-5 (101),gfs (23712),ifs (693),jcdas (336)
ensemble: r10i1p1 (420),r11i1p1 (420),r12i1p1 (420),r13i1p1 (420),r14i1p1 (420),r15i1p1 (420),r16i1p1 (420),r17i1p1 (420),r18i1p1 (420),r19i1p1 (420),r1i1p1 (2262),r1i1p1-ds4r1e8 (51),r20i1p1 (420),r21i1p1 (420),r22i1p1 (420),r23i1p1 (420),r24i1p1 (420),r25i1p1 (420),r26i1p1 (420),r27i1p1 (420),r28i1p1 (420),r29i1p1 (420),r2i1p1 (420),r30i1p1 (420),r31i1p1 (420),r32i1p1 (420),r33i1p1 (420),r34i1p1 (420),r35i1p1 (420),r36i1p1 (420),r37i1p1 (420),r38i1p1 (420),r39i1p1 (420),r3i1p1 (420),r40i1p1 (420),r41i1p1 (420),r42i1p1 (420),r43i1p1 (420),r44i1p1 (420),r45i1p1 (420),r46i1p1 (420),r47i1p1 (420),r48i1p1 (420),r49i1p1 (420),r4i1p1 (420),r50i1p1 (420),r51i1p1 (420),r52i1p1 (420),r53i1p1 (420),r54i1p1 (420),r55i1p1 (420),r56i1p1 (420),r5i1p1 (420),r6i1p1 (420),r7i1p1 (420),r8i1p1 (420),r9i1p1 (420)
You might have also seen that some facets are not relevant at all as
they are not partitioning the resulting data (e.g. see version or
grid). You can leave them out by adding the --relevant-only flag
$ freva --databrowser --all-facets --count --relevant-only data_type=reanalysis time_frequency=6hr
cmor_table: 6hr (51),6hrplev (25362)
institute: ecmwf (693),jma-criepi (336),miklip-module-c (51),nasa-gmao (101),ncep-ncar (520),noaa-cires (23712)
experiment: 20cr (23520),20cr-em (192),cfsr (96),era40 (200),era40-hr (126),eraint (367),eraint-eur-22-cclm (51),jra-25 (132),jra-25-fc (204),merra (101),ncep1 (390),ncep2 (34)
variable: cape (1),cin (1),clh (1),clivi (1),cll (1),clm (1),clt (1),clwvi (1),hur1000 (1),hur250 (1),hur500 (1),hur700 (1),hur850 (1),hur925 (1),hurs (1),hus1000 (1),hus250 (1),hus500 (1),hus700 (1),hus850 (1),hus925 (1),pr (34),prhmax (34),prw (1),psl (8169),sfcwind (1),sfcwindmax (34),sli (1),ta (238),ta1000 (1),ta250 (1),ta500 (1),ta700 (1),ta850 (1),ta925 (1),tas (101),tasmax (34),tasmin (34),td2m (1),tos (34),ua (143),ua1000 (1),ua250 (1),ua500 (1),ua700 (1),ua850 (1),ua925 (1),uas (7922),va (143),va1000 (1),va250 (1),va500 (1),va700 (1),va850 (1),va925 (1),vas (7922),wap (139),zg (383),zg1000 (1),zg250 (1),zg500 (1),zg700 (1),zg850 (1),zg925 (1)
realm: atmos (25379),ocean (34)
model: cclm-eur-22 (51),cdas (424),cfs (96),geos-5 (101),gfs (23712),ifs (693),jcdas (336)
ensemble: r10i1p1 (420),r11i1p1 (420),r12i1p1 (420),r13i1p1 (420),r14i1p1 (420),r15i1p1 (420),r16i1p1 (420),r17i1p1 (420),r18i1p1 (420),r19i1p1 (420),r1i1p1 (2262),r1i1p1-ds4r1e8 (51),r20i1p1 (420),r21i1p1 (420),r22i1p1 (420),r23i1p1 (420),r24i1p1 (420),r25i1p1 (420),r26i1p1 (420),r27i1p1 (420),r28i1p1 (420),r29i1p1 (420),r2i1p1 (420),r30i1p1 (420),r31i1p1 (420),r32i1p1 (420),r33i1p1 (420),r34i1p1 (420),r35i1p1 (420),r36i1p1 (420),r37i1p1 (420),r38i1p1 (420),r39i1p1 (420),r3i1p1 (420),r40i1p1 (420),r41i1p1 (420),r42i1p1 (420),r43i1p1 (420),r44i1p1 (420),r45i1p1 (420),r46i1p1 (420),r47i1p1 (420),r48i1p1 (420),r49i1p1 (420),r4i1p1 (420),r50i1p1 (420),r51i1p1 (420),r52i1p1 (420),r53i1p1 (420),r54i1p1 (420),r55i1p1 (420),r56i1p1 (420),r5i1p1 (420),r6i1p1 (420),r7i1p1 (420),r8i1p1 (420),r9i1p1 (420)
If you try to retrieve all variables stored (remember there are over 15 million files!) you’ll notice that some searches might bring overwhelming results:
$ freva --databrowser --facet variable
variable: abs550aer,acabf,acabfis,aerasymbnd,aeroptbnd,aerssabnd,ageice,agesno,agessc,airmass,al,albc,albdiffbnd,albdirbnd,albedo,albisccp,albsn,amo,amo-g,aoanh,aod550volso4,arag,aragos,areacella,areacello,areacellr,ares,atlantic_moc,avspsbl,bacc,baccos,baresoilfrac,basin,bbi-tslen,bbihghatm90frac,bbitm90frac,bddtalk,bddtdic,bddtdife,bddtdin,bddtdip,bddtdisi,bfe,bfeos,bigthetao,bigthetaoga,bldep,bmelt,bry,bsi,bsios,budg,budgsatm,burntarea,burntfractionall,c13land,c13litter,c13soil,c13veg,c14land,c14litter,c14soil,c14veg,c2h2,c2h6,c3h6,c3h8,c3pftfrac,c4pftfrac,calc,calcos,cape,ccb,ccldncl,ccn,cct,ccwd,cdd,cddetccdi,cdnc,cfad2lidarsr532,cfaddbze94,cfadlidarsr532,cfc11,cfc113global,cfc11global,cfc12,cfc12global,cfl,cflstddev,ch3coch3,ch4,ch4global,cheaqpso4,chegpso4,chepasoa,chepsoa,chl,chl3d,chlcalc,chlcalcos,chldiat,chldiatos,chldiaz,chldiazos,chlmisc,chlmiscos,chlos,chlpico,chlpicoos,ci,cin,cl,cland,clc,clcalipso,clcalipso2,clcalipsoice,clcalipsoliq,clccalipso,cldicemxrat,cldnci,cldncl,cldnvi,cldwatmxrat,cleaf,clh,clhcalipso,cli,clic,climodis,clis,clisccp,clitter,clitterabove,clitterbelow,clittercwd,clittergrass,clitterlut [...]
There is no upper limit for the number of facets shown. Here [...]
has been written ad hoc because for this particular case
the output comprised more than different 1,700 variable types. To avoid
that, we can stablish an upper bound number of outputs with
facet.limit. That’s the number of results that will be retrieved.
Remember that setting it to -1 retrieves just everything again… be
aware that make cause some problems if you don’t know what you are doing
(well sometimes it might also cause problems if you do… so use with
discretion)
$ freva --databrowser --facet variable facet.limit=100
variable: abs550aer,acabf,acabfis,aerasymbnd,aeroptbnd,aerssabnd,ageice,agesno,agessc,airmass,al,albc,albdiffbnd,albdirbnd,albedo,albisccp,albsn,amo,amo-g,aoanh,aod550volso4,arag,aragos,areacella,areacello,areacellr,ares,atlantic_moc,avspsbl,bacc,baccos,baresoilfrac,basin,bbi-tslen,bbihghatm90frac,bbitm90frac,bddtalk,bddtdic,bddtdife,bddtdin,bddtdip,bddtdisi,bfe,bfeos,bigthetao,bigthetaoga,bldep,bmelt,bry,bsi,bsios,budg,budgsatm,burntarea,burntfractionall,c13land,c13litter,c13soil,c13veg,c14land,c14litter,c14soil,c14veg,c2h2,c2h6,c3h6,c3h8,c3pftfrac,c4pftfrac,calc,calcos,cape,ccb,ccldncl,ccn,cct,ccwd,cdd,cddetccdi,cdnc,cfad2lidarsr532,cfaddbze94,cfadlidarsr532,cfc11,cfc113global,cfc11global,cfc12,cfc12global,cfl,cflstddev,ch3coch3,ch4,ch4global,cheaqpso4,chegpso4,chepasoa,chepsoa,chl,chl3d,chlcalc...
By the way, do you want to count them? Those are 1,756 variables!
$ freva --databrowser --facet variable | tr ',' '\n' | wc -l
1756
3. Auto-completion#
The databrowser has auto-completion embedded in it. If you are using bash, everything is already setup when you
issued the module load <freva> command. Whenever you hit tab the word
will be completed to the longest unique string that matches your
previous input. A second tab will bring up a list of all possible
completions after that.
For example ( denotes pressing the tab key):
$ freva --databrowser time_freq<TAB>
results in
$ freva --databrowser time_frequency=
Now pressing again will show all other possibilities:
$ freva --databrowser time_frequency=<TAB>
1hr 6hr amon dec grid mon monpt sem subhrpt year
3hr 6hrpt ann djf jja monc month son subyr yr
3hrpt amjjas day fx mam monclim ondjfm subhr time_series yrpt
But flags are not the only thing being populated, it also work on
attributes (Be aware that after --databrowser there is a
space):
$ freva --databrowser <TAB><TAB>
cmor_table= ensemble= grid= model= project= time_frequency=
data_type= experiment= institute= product= realm= variable=
… and of course values:
$ freva --databrowser grid=gr<TAB><TAB>
gr gr1 gr1z gr2 gr2z gr3 gra grg grz
And this is also query aware:
$ freva --databrowser institute=<TAB>
as-rcec cnes essl inm mpi-esm1-2-hr ncep-ncar preop-edf
atmos cnrm eth inpe mpi-esm-hr nerc remss
awi cnrm-cerfacs fio ipsl mpi-esm-lr nicam rmib-ugent
b1 cola-cfs fio-qlnm ipsl-cm6a-lr mpi-m nimr-kma smhi
bcc cru fub ipsl-ineris mpi-m-1 nims-kma snu
bnu csiro fub-dwd jma-criepi mpi-m-2 niwa thu
cams csiro-arccss fub-ifm knmi mpi-m-3 noaa ua
canesm5 csiro-bom geomar lasg-cess mpi-m-4 noaa-cires ub
cas csiro-qccce gerics lasg-iap mpi-m-5 noaa-gfdl [...]
$ freva --databrowser data_type=reanalysis institute=<TAB><TAB>
ecmwf jma-criepi miklip-module-c nasa-gmao ncep-ncar noaa noaa-cires zmaw
Note
If you mix flags this might not work as intended (or not at all).
4. Examples#
Apply regular expressions to the search. Find numbered
mipproducts ofcmip6, that include either meridional or zonal wind speed, or temperature (surface or at different levels). Reduce the search to certain ensemble members:
$ freva --databrowser project=cmip6 product=/.*[0-9].*/mip variable='/ta[s]*|vu/' ensemble=/r[1,3]i1p[0-9]f1/ --all-facets
...
product: c4mip,ls3mip
...
variable: ta,tas
...
ensemble: r1i1p1f1,r1i1p2f1,r3i1p1f1,r3i1p2f1
Find out all the relevant facets at official (not user crawled)
ERA5data forhourlyandmonthlytime frequencies:
$ freva --databrowser data_type=reanalysis experiment=era5 time_frequency=1hr time_frequency=mon --all-facets --count --relevant-only
cmor_table: 1hr (33166),1hr-cf (8682),1hr-era5 (6128),amon (2451),limon (86),lmon (301),mon-cf (688),mon-era5 (516),oimon (43),omon (43)
realm: atmos (40473),land (8307),landice (2216),ocean (554),seaice (554)
time_frequency: 1hr (47976),mon (4128)
variable: asn (554),bld (553),cape (553),clivi (553),clt (554),clwvi (553),cvh (554),cvl (554),esn (553),evspsbl (553),fal (554),flsr (554),fsr (554),gwd (553),hcc (554),hfls (553),hfss (553),lcc (554),lspf (553),mcc (554),mrro (553),pr (553),prcprof (553),prlsns (553),prlsprof (553),prsn (553),prsnc (553),prw (554),ps (554),psl (554),rlds (553),rldscs (553),rlut (553),rlutcs (553),rsds (553),rsdscs (553),rsn (554),rss (553),rsut (553),rsutcs (553),sic (554),skt (554),snd (554),snm (553),src (554),strd (553),swvl1 (554),swvl2 (554),swvl3 (554),swvl4 (554),ta (2765),tas (554),tasmax (553),tasmaxx (43),tasmin (553),tasminn (43),tauu (553),tauv (553),tco3 (554),tcw (554),tdps (554),tisr (553),tos (554),tsl1 (554),tsl2 (554),tsl3 (554),tsl4 (554),tsn (554),tvh (554),tvl (554),ua (2765),uas (554),uvb (553),va (2765),vas (554),wsgsmax (553),xgwdparam (553),ygwdparam (553),zg (2765),zmla (553)
ensemble: r1i1p1 (41044),r1i1p1-030000pa (2216),r1i1p1-050000pa (2216),r1i1p1-070000pa (2216),r1i1p1-085000pa (2196),r1i1p1-100000pa (2216)
Find out if a file was republished under different versions with
--multiversionfeature:
$ freva --databrowser --multiversion --facet version data_type=baseline0 variable=tas time_frequency=mon ensemble=r1i1p1
version: 20110812,20111005,20111006,20111014,20111119,20111122,20120308,20120315,20120529,20120602,20120627,20120806,20120823,20120917,20140225
Find what a given variable name stands for (e.g.
wetso4):
$ ncdump -h $(freva --databrowser variable=wetso4 | head -n 1 )| grep wetso4:standard_name
wetso4:standard_name = "tendency_of_atmosphere_mass_content_of_sulfate_expressed_as_sulfur_dry_aerosol_due_to_wet_deposition" ;
Usage in web#
The usage through web is more intuitive but less flexible as via CLI. For example, for CMIP-DICAD:
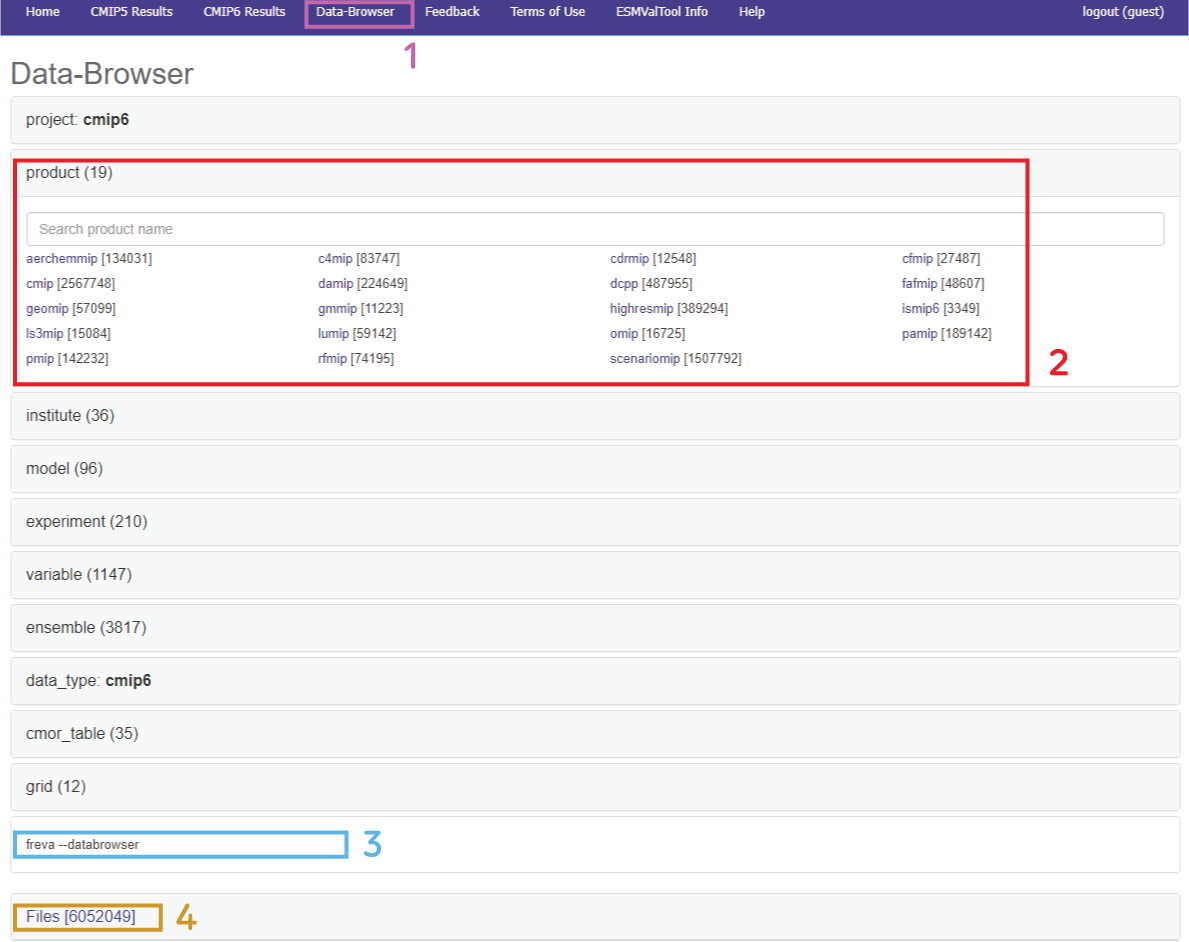
The
Data Browseris accessed clicking its corresponding tab (box 1). In this menu already the 12 facets are visible, with the number of different values for each facet in parenthesis.Every facet can be unfolded clicking on it (2), revealing all the different values contained within. Beside each value, in parenthesis, is the number of files grouped within. The values can be selected either clicking or writing them in the menu bar below the name of each facet. This text entry can be either an exact name of the value or an approximate name of a variable (e.g.
temportemperaturewould select variables such asta,tasbut alsosstetc.).The values narrow down at the path of the typed text.The instruction are translated to its CLI equivalent (
freva --databrowser [options]) ready to be copied into the terminal (3).Everytime one facet value is selected the folder tree collapses narrowing down the number of accessible total files (in 4).
Once the desired query is completed a certain amount of files will be shown. For example:
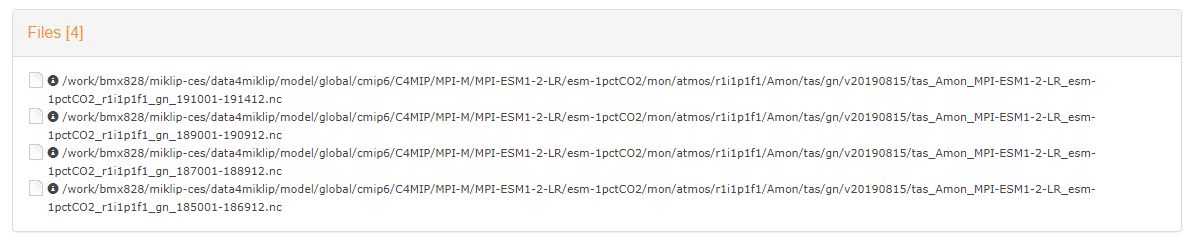
would correspond to the following query:
$ freva --databrowser project=cmip6 product=c4mip institute=mpi-m experiment=esm-1pctco2 time_frequency=mon realm=atmos variable=tas
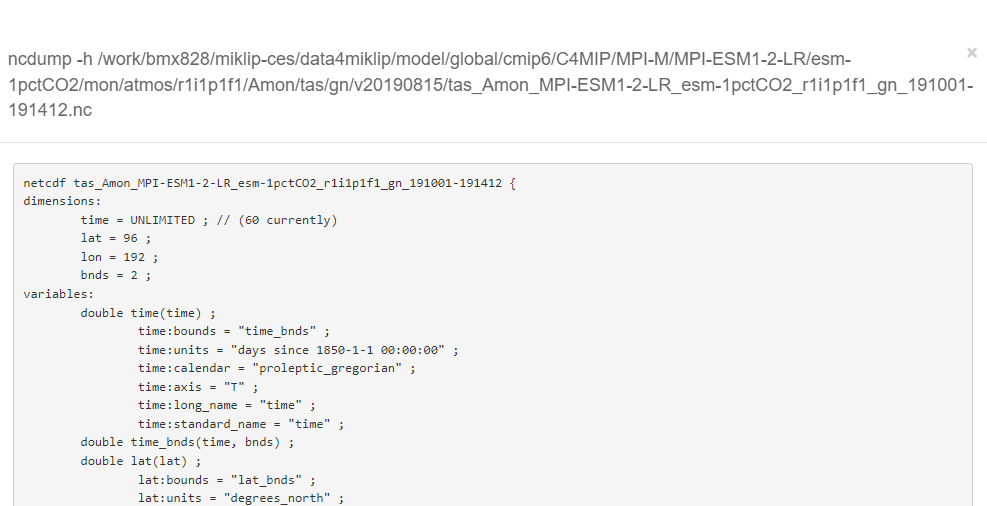
Freva allows the inspection of the metadata of each file. For that it runs a ncdump -h over the file once the information icon is clicked.
The execution of the command is done via ssh on the credentials of the user, hence, it asks for the LDAP password to complete it (only the first time):
Note
Please be aware that path the SOLR server shows on the data browser only reflects the state of the files at the time of the snapshot. It does not necessarily correspond to a currently existing file anymore, as the file could have been either renamed, moved or erased after the indexation took place.